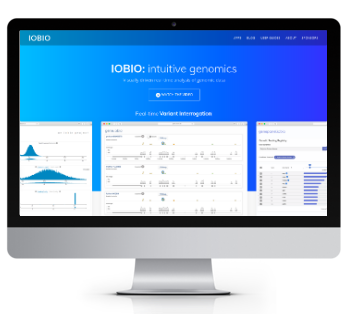
iobio
Realtime genomic data visualization and analysis web tools
- gene.iobio - interactive visual variant annotation and prioritization
- clin.iobio - comprehensive clinical variant review workflow
- genepanel.iobio - phenotype and disorder associated gene lists
- bam.iobio - alignment file quality control
- vcf.iobio - variant calling file quality control
- oncogene.iobio - variant annotation and tracking in oncology studies
- taxonomer.iobio - ultra-fast metagenomic analysis